#include <spikingneuron.h>
Public Member Functions | |
| SpikingNeuron (void) | |
| virtual | ~SpikingNeuron (void) |
| virtual string | name (void) const |
| virtual int | dimension (void) const =0 |
| virtual void | variables (vector< string > &varnames) const =0 |
| virtual void | units (vector< string > &u) const =0 |
| virtual void | operator() (double t, double s, double *x, double *dxdt, int n)=0 |
| virtual void | init (double *x) const =0 |
| virtual void | conductances (vector< string > &conductancenames) const |
| virtual void | conductances (double *g) const |
| virtual string | conductanceUnit (void) const |
| virtual void | currents (vector< string > ¤tnames) const |
| virtual void | currents (double *c) const |
| virtual string | currentUnit (void) const |
| virtual string | inputUnit (void) const |
| virtual void | add (void) |
| virtual void | notify (void) |
| double | gain (void) const |
| double | offset (void) const |
 Public Member Functions inherited from ConfigClass Public Member Functions inherited from ConfigClass | |
| ConfigClass (const string &ident, int group=0, int mode=0xffff, int selectmask=0) | |
| ConfigClass (const ConfigClass &C) | |
| virtual | ~ConfigClass (void) |
| const string & | configIdent (void) const |
| void | setConfigIdent (const string &ident) |
| int | configGroup (void) const |
| void | setConfigGroup (int group) |
| int | configMode (void) const |
| void | setConfigMode (int mode) |
| void | addConfigMode (int mode) |
| void | delConfigMode (int mode) |
| int | configSelectMask (void) const |
| void | setConfigSelectMask (int mask) |
| void | addConfig (void) |
| void | readConfig (void) |
| virtual void | readConfig (StrQueue &sq) |
| virtual void | saveConfig (ofstream &str) |
| virtual int | configSize (void) const |
| virtual void | config (void) |
| virtual void | preConfig (void) |
Static Public Attributes | |
| static const int | ScalingFlag = 16 |
| static const int | ModelFlag = 32 |
| static const int | DescriptionFlag = 64 |
Protected Attributes | |
| double | Gain |
| double | Offset |
Additional Inherited Members | |
 Public Types inherited from ConfigClass Public Types inherited from ConfigClass | |
| enum | Action |
 Static Public Member Functions inherited from ConfigClass Static Public Member Functions inherited from ConfigClass | |
| static void | setConfigureClasses (ConfigureClasses *cfg) |
Detailed Description
[ModelLib] Base class for a spiking (point-) neuron
- Version
- 1.2 (May 7, 2008)
- SpikingNeuron inherits Options
- Added interface for accessing the values of the ionic currents and conductances
- 1.1 (Jan 10, 2006)
Each model of a spiking neuron has a name(). The model is implemented as a set of differential equations
![\[ \left( \begin{array}{c} \frac{dx_1}{dt} \\ \frac{dx_2}{dt} \\ \vdots \\ \frac{dx_n}{dt} \end{array} \right) = \left( \begin{array}{c} f_1(x_1, x_2, \ldots, x_n, s) \\ f_2(x_1, x_2, \ldots, x_n, s) \\ \vdots \\ f_n(x_1, x_2, \ldots, x_n, s) \end{array} \right) \]](form_0.png)
The state of the model is described by the state vector 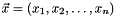 of dimension n = dimension(). operator()() computes the derivatives
of dimension n = dimension(). operator()() computes the derivatives 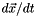 with respect to time t for the current state x and the stimulus s. The variables() function returns names for each of the state variables x, units() returns the corrsponding units, and init() sets the state variables x to useful initial conditions. The unit of the input s is given by inputUnit().
with respect to time t for the current state x and the stimulus s. The variables() function returns names for each of the state variables x, units() returns the corrsponding units, and init() sets the state variables x to useful initial conditions. The unit of the input s is given by inputUnit().
While integrating the model, the current values of the ionic currents and their corresponding conductances can be retrieved by the currents(double*) const and conductances(double*) const functions. The corresponding names of the currents and conductances are returned by conductances(vector<string>&) const and currents(vector<string>&) const, respectively. The unit of the conductances is conductanceUnit() and the the one of the currents is currentUnit().
Parameter values of the model can be made accessible by adding them to the Options in add(). Changed parameter values are read out from the Options by notify(). The parameter values are classified as either scalingFlag(), modelFlag(), or descriptionFlag().
SpikingNeuron defines two parameters offset() and gain() with default values 0 and 1, respectively, that should be applied to whatever input before it is passed on as the stimulus s for computing the derivatives via operator()().
Constructor & Destructor Documentation
| SpikingNeuron | ( | void | ) |
Constructs the model, i.e. initializes model parameter with useful default values.
- Note
- The constructor should not add any new options. This goes into the add() function.
References SpikingNeuron::name(), and ConfigClass::setConfigIdent().
|
virtual |
Member Function Documentation
|
virtual |
- Returns
- the name of the model.
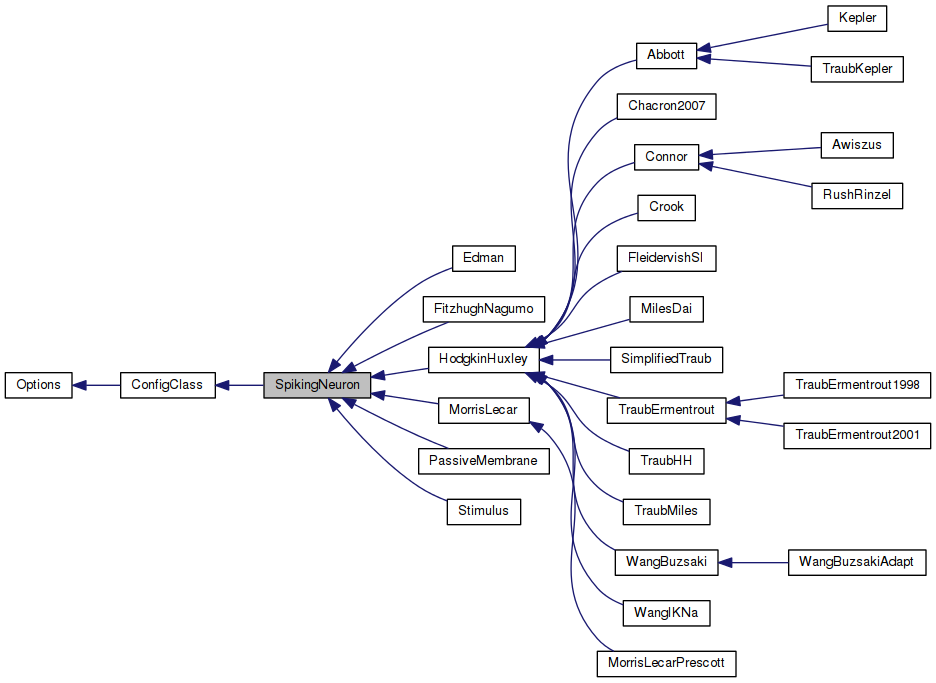
Reimplemented in Chacron2007, Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, WangBuzsaki, SimplifiedTraub, TraubErmentrout2001, TraubErmentrout1998, TraubKepler, TraubMiles, TraubHH, FleidervishSI, Awiszus, RushRinzel, Connor, Kepler, Abbott, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, PassiveMembrane, and Stimulus.
Referenced by NeuronModels::add(), NeuronModels::metaData(), and SpikingNeuron::SpikingNeuron().
|
pure virtual |
- Returns
- the dimension of the system.
- See Also
- variables(), operator()()
Implemented in Chacron2007, Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, WangBuzsaki, SimplifiedTraub, TraubErmentrout, TraubMiles, FleidervishSI, RushRinzel, Connor, Abbott, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, PassiveMembrane, and Stimulus.
Referenced by NeuronModels::main().
|
pure virtual |
- Parameters
-
[out] varnames the names of each of the dimension() variables. Gating variables (ranging between 0 and 1) should be a single lower-case character, potentials and equivalent potentials a single upper-case character, and concentrations should be in brackets (e.g. [Ca]).
- See Also
- dimension(), units(), operator()()
Implemented in Chacron2007, Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, WangBuzsaki, SimplifiedTraub, TraubErmentrout, TraubMiles, FleidervishSI, RushRinzel, Connor, Abbott, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, PassiveMembrane, and Stimulus.
|
pure virtual |
- Parameters
-
[out] u the units of the variables returned by variables().
- See Also
- dimension(), operator()()
Implemented in Chacron2007, Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, WangBuzsaki, SimplifiedTraub, TraubErmentrout, TraubMiles, FleidervishSI, RushRinzel, Connor, Abbott, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, PassiveMembrane, and Stimulus.
|
pure virtual |
Computes the derivative dxdt at time t with stimulus s given the state x. Implement this function with your model.
- Parameters
-
[in] t the time. [in] s the stimulus. [in,out] x the state vector. [out] dxdt the derivative with respect to time. [in] n the number of variables, usually equal to dimension().
- See Also
- dimension(), init(), variables(), inputUnit()
Implemented in Chacron2007, Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, WangBuzsaki, SimplifiedTraub, TraubErmentrout2001, TraubErmentrout1998, TraubKepler, TraubMiles, TraubHH, FleidervishSI, Awiszus, RushRinzel, Connor, Kepler, Abbott, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, PassiveMembrane, and Stimulus.
|
pure virtual |
Initialize the state x with useful inital conditions.
- Parameters
-
[out] x the dimension() state variables of the model.
- See Also
- dimension(), operator()()
Implemented in Chacron2007, Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, WangBuzsaki, SimplifiedTraub, TraubErmentrout2001, TraubErmentrout1998, TraubKepler, TraubMiles, TraubHH, FleidervishSI, Awiszus, RushRinzel, Connor, Abbott, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, PassiveMembrane, and Stimulus.
Referenced by NeuronModels::main().
|
virtual |
Implement this function to return the names of the individual ionic conductances that conductances(double*) const would return. The default implementation returns an empty vector.
- Parameters
-
[out] conductancenames the names of the individual ionic conductances.
- See Also
- currents(double*) const
Reimplemented in Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, TraubErmentrout, TraubMiles, Connor, HodgkinHuxley, MorrisLecarPrescott, and MorrisLecar.
|
virtual |
Implement this function to return in g the values of the individual ionic conductances. The number of conductances is defined by the size of conductancenames the function conductances(vector<string>&) const returns. The default implementation returns nothing.
- Parameters
-
[out] g the current values of the individual ionic conductances.
- See Also
- currents(double*) const
Reimplemented in Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, TraubErmentrout, TraubMiles, Connor, HodgkinHuxley, MorrisLecarPrescott, and MorrisLecar.
|
virtual |
Returns the unit of the conductances returned by conductances(double*) const. The default implementation returns mS/cm^2.
Reimplemented in Chacron2007, Edman, MilesDai, and MorrisLecar.
Referenced by HodgkinHuxley::add(), Connor::add(), TraubMiles::add(), TraubErmentrout::add(), WangBuzsakiAdapt::add(), Crook::add(), and WangIKNa::add().
|
virtual |
Implement this function to return in currentnames the names of the individual ionic currents that currents(double*) const would return. The default implementation returns an empty vector.
- Parameters
-
[out] currentnames the names of the individual ionic currents.
- See Also
- conductances(double*) const
Reimplemented in Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, TraubErmentrout, TraubMiles, Connor, HodgkinHuxley, MorrisLecarPrescott, and MorrisLecar.
|
virtual |
Implement this function to return in c the values of the individual ionic currents. The number of currents is defined by the size of currentnames the function currents(vector<string>&) const returns. The default implementation returns nothing.
- Parameters
-
[out] c the values of the individual ionic currents.
- See Also
- conductances(double*) const
Reimplemented in Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, TraubErmentrout, TraubMiles, Connor, HodgkinHuxley, MorrisLecarPrescott, and MorrisLecar.
|
virtual |
Returns the unit of the currents returned by currents(double*) const. The default implementation returns uA/cm^2.
Reimplemented in Chacron2007, Edman, MilesDai, and MorrisLecar.
|
virtual |
Returns the unit of the input (the stimulus). The default implementation returns uA/cm^2.
- See Also
- operator()()
Reimplemented in Chacron2007, Edman, MilesDai, and MorrisLecar.
|
virtual |
Implement this function to add all necessary options. Set the flags of each option to either one of ScalingFlag, ModelFlag, or DescriptionFlag. For example:
This implementation adds a label "Input" followed by two numbers "Gain" and "Offset". Call it in your reimplementation via
- See Also
- notify()
Reimplemented in Chacron2007, Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, SimplifiedTraub, TraubErmentrout1998, TraubErmentrout, TraubMiles, RushRinzel, Connor, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, and PassiveMembrane.
References SpikingNeuron::Gain, SpikingNeuron::Offset, and SpikingNeuron::ScalingFlag.
Referenced by NeuronModels::add(), PassiveMembrane::add(), FitzhughNagumo::add(), MorrisLecar::add(), HodgkinHuxley::add(), Crook::add(), MilesDai::add(), WangIKNa::add(), and Edman::add().
|
virtual |
Implement this function to read out the current values from the list of Options. For example:
This implementation reads out the gain() and the offset(). Call it in your reimplementation via
- See Also
- add()
Reimplemented in Edman, WangIKNa, MilesDai, Crook, WangBuzsakiAdapt, SimplifiedTraub, TraubErmentrout1998, TraubErmentrout, TraubMiles, RushRinzel, Connor, HodgkinHuxley, MorrisLecarPrescott, MorrisLecar, FitzhughNagumo, and PassiveMembrane.
References SpikingNeuron::Gain, and SpikingNeuron::Offset.
Referenced by PassiveMembrane::notify(), FitzhughNagumo::notify(), MorrisLecar::notify(), MorrisLecarPrescott::notify(), HodgkinHuxley::notify(), Connor::notify(), RushRinzel::notify(), Crook::notify(), MilesDai::notify(), WangIKNa::notify(), Edman::notify(), and NeuronModels::readOptions().
| double gain | ( | void | ) | const |
- Returns
- the gain that should be applied to the input.
- See Also
- offset()
References SpikingNeuron::Gain.
Referenced by NeuronModels::operator()().
| double offset | ( | void | ) | const |
- Returns
- the offset that should be applied to the input.
- See Also
- gain()
References SpikingNeuron::Offset.
Referenced by NeuronModels::operator()().
Member Data Documentation
|
static |
Flag for selecting input / output gain and offset options.
Referenced by SpikingNeuron::add(), FitzhughNagumo::add(), and MorrisLecar::add().
|
static |
Flag for selecting the model options.
Referenced by PassiveMembrane::add(), FitzhughNagumo::add(), MorrisLecar::add(), MorrisLecarPrescott::add(), HodgkinHuxley::add(), Connor::add(), RushRinzel::add(), TraubMiles::add(), TraubErmentrout::add(), TraubErmentrout1998::add(), SimplifiedTraub::add(), WangBuzsakiAdapt::add(), Crook::add(), MilesDai::add(), WangIKNa::add(), and Edman::add().
|
static |
Flag for selecting the model description.
|
protected |
The gain that should be applied to the input.
Referenced by SpikingNeuron::add(), SpikingNeuron::gain(), MorrisLecar::MorrisLecar(), MorrisLecarPrescott::MorrisLecarPrescott(), and SpikingNeuron::notify().
|
protected |
The offset that should be applied to the input.
Referenced by SpikingNeuron::add(), MorrisLecar::MorrisLecar(), MorrisLecarPrescott::MorrisLecarPrescott(), SpikingNeuron::notify(), and SpikingNeuron::offset().
The documentation for this class was generated from the following files:
 1.8.6
1.8.6